Improving on Nature’s Design: Possibilities and Implementation
There are two parts to that project. The first deals with the foundations of energy transformations performed by Life and with the question of how available free-energy input can be optimally channeled toward Life’s preservation and multiplication. The second deals with the discovery and rational design of peptides that could interact with biological membranes in a manner beneficial to medical treatments.

Figure 1. The cover page for D. Juretić book on bioenergetics (2021 E-book and 2022 hardcover).
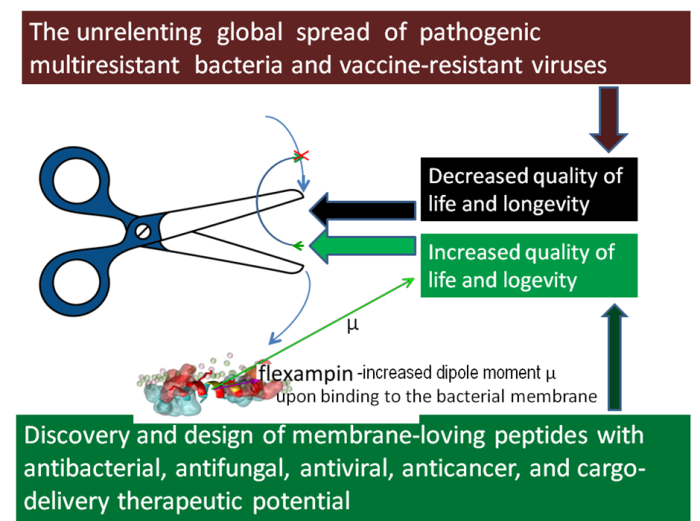
Figure 2. A concept illustration for the book-writing plan about membrane-loving enzymes.
Short project description
My just-published book on bioenergetics (Figure 1) summarizes my lifelong search for answers to how rivers of Life forever change Life and its environment by coupling biological to thermodynamic evolution. Life scientists can speed up both evolutions by rational design of peptides and proteins. The first project goal is to optimize nanocurrents produced by enzymes by the requirement of maximal dissipation in rate-limiting catalytic steps. The second goal is to increase the availability of novel membrane-loving peptides I discovered or designed with possible therapeutic applications. That will be the topic of a book I am now writing (Figure 2).
Work in 2021 year / Introduction / Work description and results
The rational way to optimize the efficiency of biochemical catalytic cycles requires a better understanding of Life from an irreversible thermodynamics viewpoint. My monograph on bioenergetics (Juretić, 2021) offers the following definition. Life is the process of decreasing available forces Xa through primary fluxes Ja and secondary fluxes Js. It results mainly in additional dissipation and partially into charge separation, chemical transformations, and the accumulation of power-producing secondary force Xs times Js couples XsJs for self-maintenance and reproduction – the processes that increase the complexity of Life and the entropy of Life's environment. It is a physicist's viewpoint, with an emphasis on power couples XsJs that contribute negative terms to total entropy production, thus increasing the efficiency of free-energy transduction without breaking the Second Law of thermodynamics.
The book (Juretić, 2021) summarizes and extends previously published interdiciplinary results of D. Juretić and his collaborators scattered through years in scientific journals and books belonging to different research fields (Dobovišek et al., 2014; Bonačić Lošić et al. 2017; Juretić et al. 2019a, 2019b; Juretić and Bonačić Lošić 2021; Juretić 2021). It also ventures into a re-evaluation of under-appreciated ideas of Erwin Schrödinger, Peter Mitchell, Ilya Prigogine, Terrel Hill, and other authors. The book explores the usefulness of dissecting the entropy production of enzymes involved in cellular defenses, fermentation, respiration, and photosynthesis, assuming that tightly regulated dissipation is the hallmark of Life. It stresses the need for an interdisciplinary approach to study cells' virtuoso ability to connect the microscopic to the macroscopic world.
The book also explores optimization principles proposed through the years for biomolecular machines. When applied to bioenergetics and enzyme kinetics, we found unrealistic aspects of both minimal and maximal principles for total entropy production. Neither does justice to strict regulation of overall entropy production level, which is obligatory for the survival of a cell or organism in an often hostile environment. A natural tendency for maximal possible increase of entropy production is allowed by the cell to take its course only in catalytic steps subjugated to a biological need to preserve and multiply its complex, low probability structures. Thus, we should maximize partial entropy production in rate-limiting steps to increase an overall entropy production instead of maximizing total entropy production. As a pedagogical example, we published the Comment (Juretić and Bonačić Lošić, 2021) with a critical appraisal of forcing the triosephosphate isomerase enzyme (TPI) to have a well-defined maximum in overall entropy production and information entropy. TPI catalyzes the conversion from DHAP to GAP, which is subsequently used by downstream glycolytic enzymes. It is a reversible enzyme in the sense that it works close to equilibrium in physiological conditions and can be easily controlled to synthesize either GAP or DHAP according to cell needs, but always with a low amount of associated overall entropy production (Bonačić Lošić et al., 2017) Simulations performed by Andrej Dobovišek and his co-authors (Šterk et al., 2021) led to TPI thermodynamic and biological dysfunction with an associated 600 times higher overall entropy production. Their optimization of the TPI enzyme converted that reversible enzyme into an irreversible monster engine for inhibiting the glycolysis and creating an excess of toxic DHAP decomposition products. A take-home message is that a natural desire of theoreticians to simplify things should not have precedence over biological and biochemical reality.
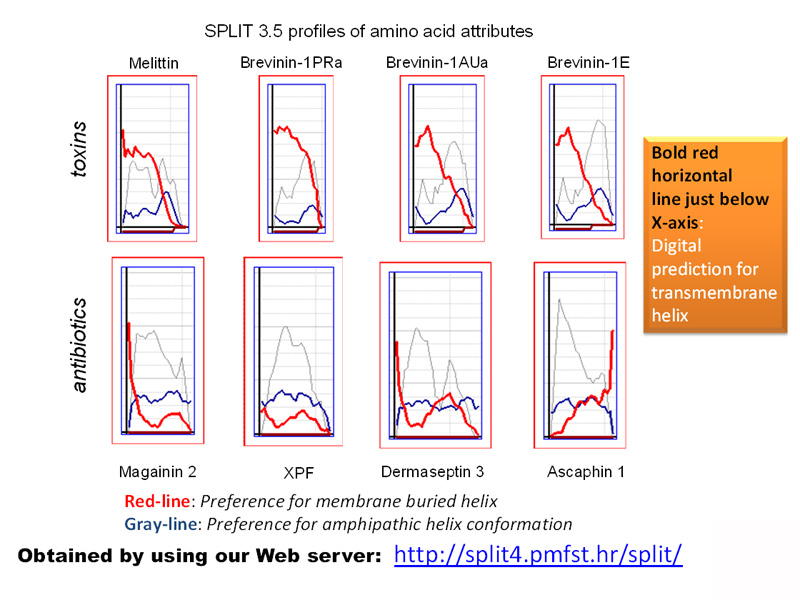
Book-writing about membrane-loving peptides turned out to be explorations within the second part of the project. The initial idea was about disseminating simple bioinformatic tools and rational approaches to discovering and designing novel peptide antibiotics with confirmed predictions of high antimicrobial activity and selectivity by our home-constructed algorithms and web servers (Juretić et al., 2011; Tessera et al., 2012; Kamech et al., 2012, Novković et al., 2012; Ilić et al., 2013, Kozić et al., 2015; Juretić et al., 2015; Rončević et al., 2017; Juretić et al., 2017; Juretić et al. 2018, Juretić and Simunić 2019; Juretić et al., 2020). It morphed when confronted with the multifunctional nature of most natural lead peptides. Cases of simultaneous antibacterial, antifungal, antiviral, and anticancer activity are anything but rare. Interaction of cationic peptides with anionic membrane surface is only one face of a complex story on how soluble peptides attach to the membrane, transfer through it, and bind to intracellular targets such as DNA. For cell-penetrating peptides, membrane transfer is “painless” for human membranes, even when such peptides carry a bulky cargo molecule that can have some beneficial therapeutic effect. Hybrid peptides and peptide conjugates are human creations with vast possibilities for applications in medicine and biotechnology. Their advantage is often the precision targeting body cells or organs in a need of treatment and an absence of bacterial or body resistance to such drugs. The search for such natural or designed lead compounds benefits from interdisciplinary research connecting bioinformatics, biophysics, and physics. For instance, we found several years ago that designed peptide flexampin undergoes a huge increase in its dipole moment μ after attachment to anionic (bacterial-like) membranes (see the second figure). That observation helped to understand the superior antibacterial and anticancer activity and selectivity of flexampin (Juretić et al., 2017; Juretić et al., 2020).
Conclusion (including work plan for the next year)
There are two opposing approaches on how to apply entropy production principles to enzyme kinetics and bioenergetics. The first one simulates the working cycles of an enzyme by requiring an absolute maximum in total entropy production regardless of physiological direction and observed safe working range. The second one selects rate-limiting catalytic steps and maximizes partial entropy production associated with these biologically productive steps. Core chapters of my monograph on bioenergetics argue for a tight connection among increased overall entropy production, increased turnover number (catalytic constant), and increased enzyme efficiency after crucial catalytic steps have been optimized for transitional (step-specific) maximal entropy production.
We show in the book that biological evolution nearly achieved such optimization for enzymes performing the free-energy transduction. For simpler substrate to product converting enzymes, additional catalytic improvements can be achieved, despite biochemical description for some of them as “perfect enzymes.” One such example is the housekeeping enzyme Triosephosphate isomerase. In the book and the Comment published this year we show that our approach speeds up TPI product formation which is vital for glycolysis, while brute-force MaxEP application (the first approach) does not.
During the following year (2022), we intend to get additional results supporting our viewpoint that Life strives to regulate and increase its total entropy production by allowing for near-maximal partial entropy production only in rate-limiting catalytic steps. The guide for these simulations on other housekeeping enzymes will be our theorem about maximal partial entropy production (Juretić et al., 2019). For the second part of the project (membrane-loving peptides), I expect to finish the book-writing job in 2022. Since book-writing is accompanied by data mining and applications of numerous predictive algorithms, the results will also be promising sequences of natural and designed peptides with a wide spectrum of possible applications.
References
- Željana Bonačić Lošić, Tomislav Donđivić and Davor Juretić. Is the catalytic activity of triosephosphate isomerase fully optimized? An investigation based on maximization of entropy production. J Biol Phys. 43 (2017), 69-86. doi: 10.1007/s10867-016-9434-3 . Epub 2017 Jan 3. PubMed PMID: 28050739; PubMed Central PMCID: PMC5323346.
- Andrej Dobovišek, Paško Županović, Milan Brumen and Davor Juretić: "Maximum entropy production and maximum Shannon entropy as the germane principles for the evolution of enzyme kinetics". Published as a book chapter in Beyond the Second Law: Entropy Production and Non-Equilibrium Systems (R.C. Dewar, C. Lineweaver, R. Niven, K. Regenauer-Lieb, eds.), pp. 361-382, Springer (2014). doi: 10.1007/978-3-642-40154-1_19
- Davor Juretić, Damir Vukičević, Dražen Petrov, Mario Novković, Viktor Bojović, Bono Lučić, Nada Ilić and Alessandro Tossi. Knowledge-based computational methods for identifying or designing novel, non-homologous antimicrobial peptides. Eur. Biophys. J., 40 (2011), 371-385. doi: 10.1007/s00249-011-0674-7
- Davor Juretić, Damir Vukičević and Alessandro Tossi. Tools for Designing Amphipathic Helical Antimicrobial Peptides. Methods Mol Biol. 1548 (2017), 23-34. doi: 10.1007/978-1-4939-6737-7_2. PubMed PMID: 28013494.
- D. Juretić, Y. Sonavane, N. Ilić, G. Gajski, I. Goić-Barišić, M. Tonkić, M. Kozic, A. Maravić, F.-X. Pellay and L. Zoranić. Designed peptide with a flexible central motif from ranatuerins adapts its conformation to bacterial membranes. Biochim Biophys Acta Biomembr. 1860 (2018), 2655-2668. doi: 10.1016/j.bbamem.2018.10.005. Epub 2018 Oct 4. PubMed PMID: 30292398
- Davor Juretić, Juraj Simunić and Želana Bonačić Lošić. The maximum entropy production requirement for proton transfers enhances catalytic efficiency for β-lactamases. Biophys Chem. 244 (2019a), 11-21. doi: 10.1016/j.bpc.2018.10.004. Epub 2018 Oct 17. PubMed PMID: 30448627.
- Davor Juretić, Juraj Simunić, and Željana Bonačić Lošić. Maximum Entropy Production Theorem for Transitions between Enzyme Functional States and Its Applications. Entropy 21 (2019b), 743; doi: 10.3390/e21080743.
- Davor Juretić and Juraj Simunić. Design of α-helical antimicrobial peptides with a high selectivity index. Expert Opin Drug Discov. (2019) Jul 16:1-11. doi: 10.1080/17460441.2019.1642322. [Epub ahead of print] PubMed PMID: 31311351.
- Davor Juretić, Anja Golemac, Denise E. Strand, Keshi Chung, Nada Ilić, Ivana Goić-Barišić and François-Xavier Pellay. The Spectrum of Design Solutions for Improving the Activity-Selectivity Product of Peptide Antibiotics against Multidrug-Resistant Bacteria and Prostate Cancer PC-3 Cells. Molecules 25 (2020): 3526. doi: 10.3390/molecules25153526. PMID: 32752241; PMCID: PMC7436000.
- Davor Juretić and Željana Bonačić Lošić. Comments on ‘Flexibility of enzymatic transitions as a hallmark of optimized enzyme steady-state kinetics and thermodynamics’. Comput. Biol. Chem. 95 (2021) Sep 8: 107571. doi: 10.1016/j.compbiolchem.2021.107571.
- Davor Juretić: “Bioenergetics: A Bridge Across Life and Universe.“, Available December 23, 2021, CRC Press Taylor & Francis Group (2022), Boca Raton, Florida, USA.
- Nada Ilić, Mario Novković, Filomena Guida, Daniela Xhindoli, Monica Benincasa, Alessandro Tossi and Davor Juretić. Selective antimicrobial activity and mode of action of adepantins, glycine-rich peptide antibiotics based on anuran antimicrobial peptide sequences. BBA-Biomembranes 1828 (2013), 1004-1012. Available online from 26. November 2012. doi: 10.1016/j.bbamem.2012.11.017
- Nédia Kamech, Damir Vukičević, Ali Ladram, Christophe Piesse, Julie Vasseur, Viktor Bojović, Juraj Simunić and Davor Juretić. Improving the selectivity of antimicrobial peptides from anuran skin. Journal of Chemical Information and Modeling, 52 (2012), 3341–3351. Available online from 24. October 2012. doi: 10.1021/ci300328y
- Mara Kozić, Damir Vukičević, Juraj Simunić, Tomislav Rončević, Nikolinka Antcheva, Alessandro Tossi and Davor Juretić. Predicting the Minimal Inhibitory Concentration for Antimicrobial Peptides with Rana-box Domain. Journal of Chemical Information and Modeling 55 (2015), 2275-2287. doi: 10.1021/acs.jcim.5b00161. Epub 2015 Sep 18.
- Mario Novković, Juraj Simunić, Viktor Bojović, Alessandro Tossi and Davor Juretić. DADP: the Database of Anuran Defense Peptides. Bioinformatics J., 28 (2012), 1406-1407.
- Šterk, M., Markovič, R., Marhl, M., Fajmut, A. and Dobovišek, A. Flexibility of enzymatic transitions as a hallmark of optimized enzyme steady-state kinetics and thermodynamics. Comput. Biol. Chem. 91 (2021), 107449. doi: 10.1016/j.compbiolchem.2021.107449
- Tomislav Rončević, Goran Gajski, Nada Ilić, Ivana Goić-Barišić, Marija Tonkić, Larisa Zoranić, Juraj Simunić, Monica Benincasa, Marijana Mijaković , Alessandro Tossi and Davor Juretić. PGLa-H tandem-repeat peptides active against multidrug resistant clinical bacterial isolates. Biochimica et Biophysica Acta 1859 (2017), 228–237. doi: 10.1016/j.bbamem.2016.11.011
- Valentina Tessera, Filomena Guida, Davor Juretić and Alessandro Tossi. Identification of antimicrobial peptides from teleosts and anurans in expressed sequence tag databases using conserved signal sequences. FEBS Journal, 279 (2012), 724–736. doi: 10.1111/j.1742-4658.2011.08463.x
Antimicrobial peptides (AMPs) research
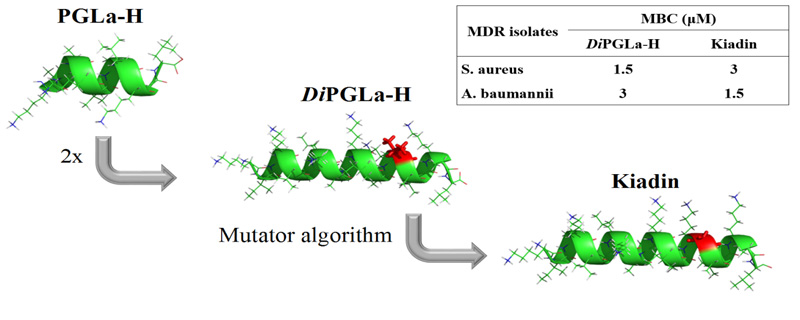
Kiadin – peptide antibiotic I designed and we examined in Split, Zagreb, Trieste and Melbourne. It is active against multidrug resistant (MDR) clinical isolates, in particular A. Baumannii, and nontoxic for circulating human blood cells. Rončević et al., Biochimica et Biophysica Acta 1859 (2017) 228–237.
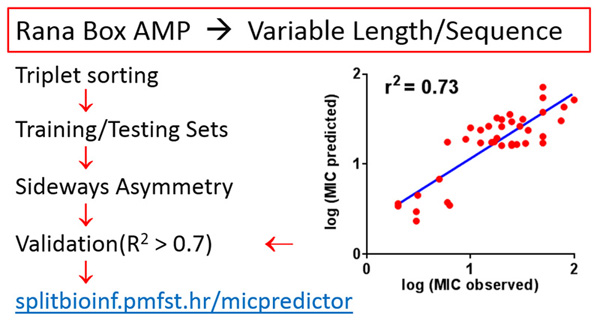
Predicting minimal inhibitory concentration (MIC) against E. coli for Rana-box antimicrobial peptides – MIC is notoriously difficult to predict, but even a modest success with that goal can save time, labour and expense in wandering without guidance through enormous space of possible antimicrobial peptide sequences. We used only the generalization of sequence moment concept, named the sideways asymmetry moment, as the key descriptor, and provided structural insight how different segments of peptide contribute to its bacteriostatic activity. We designed Ranaboxin-1 using our MIC prediction algorithm (http://splitbioinf.pmfst.hr/micpredictor/) and Kozić et al. paper: J Chem Inf Model. 2015,55, 2275-2287. It has MIC of 2 µM against Gram-negatives: E. coli, A. baumannii and P. Aeruginosa, but its predicted MIC was 0.1 µM. It is the result of the first QSAR model capable of MIC prediction for AMPs of widely different lengths and low identity, by using only one descriptor. Mara Kozić got her MSc degree from the University of Split, Faculty of Science, biophysics graduate programme for giving a major input to this research.
Adepantins – peptide antibiotics we designed at the University of Split, by using anuran antimicrobial peptides, are less than 50% identical to any other antibiotics of that type and exhibit very high selectivity for Gram-negative bacterial cells such as E. coli without being harmful to human cells (Juretić et al. 2009 and 2011). Their dimmers have minimal inhibitory concentration (MIC) of only 0.5 µM, while monomers, such as Adepantin 2, have selectivity index (SI) of about 400, which is about four times higher than the SI of the best natural peptide antibiotic isolated from frog and toads skin (Ilić et al. 2013). High selectivity index means low toxicity for human red blood cells. Nada Ilić got her PhD from the biophysics PhD programme (http://split.pmfst.unist.hr/biophysics/) for her experiments with adepantins, while D. Juretić and D. Vukičević constructed QSAR model for designing adepantins, which contained just one (but novel) descriptor.
Human-fish peptide found by myself in the database of human expressed sequence tags, is not fish AMP “living” in human genome. Its origin remains mysterious, but its close relationship with putative sticklefish AMP is quite obvious. It is possible that some technician in the USA enjoyed its fishburger, while working on decoding human transcriptome, so that fish RNA entered by accident into the reaction mixture. Anyway, experiments by Tessera et al. (2012) determined that it is a broad spectrum peptide antibiotic equally active (MIC = 1 µM) against Gram-negatives (E. coli) and Gram-positives (S. aureus), while its SI is close to 100, which is very good for any AMP. Valentina Tessera got her MSc for her experiments with the hfp.
DADP: the Database of Anuran Defense Peptides http://split4.pmfst.hr/dadp/ – published in the Bioinformatics J. (Novković et al., 2012), quickly become a useful resource for analysing the origin of anuran antimicrobial peptides and for constructing structure-activity models by using activity and toxicity data (for almost 1000 peptides) previously scattered in literature. Mario Novković got his MSc for this work from the University of Split, Faculty of Science, biophysics graduate programme.
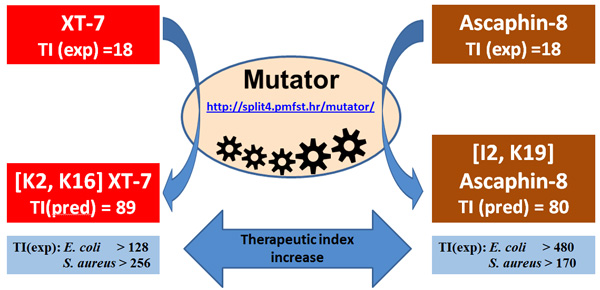
Mutator algorithm from the Kamech et al. 2012 paper (http://split4.pmfst.hr/mutator) can quickly find amino acid substitutions expected to increase selectivity index of natural antimicrobial peptides from frogs and toads (the selectivity index is named therapeutic index in that paper). When used wisely (Juretić et al., 2016) it can increase antimicrobial activity as well.
Trichoplanin – peptide antibiotic we found in the database of Trichoplax adhaerens expressed sequence tags (Simunic et al., 2014):
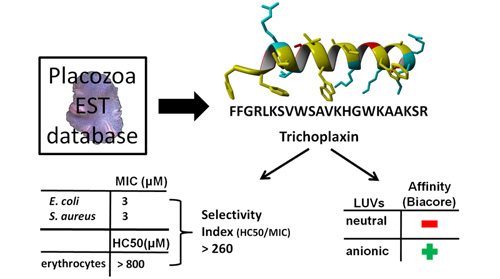
It is broad spectrum antibiotic with very high selectivity index not similar to anything known (the E-value is 10 or >10 after BLASTP search). The importance of finding it in T. adhaerens is due to possibility that T. adhaerens may be the first animal with smallest genome, and also belonging to rare species which defeated ageing and death. When dispersed into individual cells they find each other again and form the whole organism. Currently, we are examining several additional candidates for T. adhaerens antibiotics, as well as mechanism of action of peptide antibiotics and possible synergy between them, the research topics where I collected a large number of citations in an earlier period (275 and 183 respectively from 1989-1995).
Maximum entropy production (MEP) principle and its applications in physics, bioenergetics and enzyme kinetics
The MEP and maximum information entropy (MaxEnt) principles are good candidates for unique physical selection principles that act in concert with biological selection and evolution, not because we love them, but because we find better agreement with experiments in the case of enzyme kinetics (Dobovišek et al. 2011, Lošić et al. 2016) and bioenergetics (Juretić and Županović, 2003, Dewar et al. 2006, Dobovišek et al. 2014, Lošić et al., 2016). Each of proposed alternative optimization principles, such as the maximal metabolic flux, or Prigogine’s principle of minimal entropy production has serious drawbacks. Domagoj Kuić from Faculty of Science, University of Split, got his PhD by exploring MEP and MaxEnt principles within predictive statistical mechanics (Kuić et al., 2012).
For biologists, biophysicists and physicists interested in quantitative biology and in thinking about questions “What is Life” or “Why life exists in the universe” I shall cite several concluding sentences from our manuscripts during the last decade(Juretić and Županović, 2005; Lošić et al. 2016):
“Chemical and biological evolution of macromolecular structures facilitated its integration with thermodynamic evolution in a way which can be best described as a synergistic relationship or positive feedback”.
Living entities tend to increase the entropy production in the universe while active metabolically, so that life serves as a catalytic agent speeding entropy production in its environment. This observation brings biological evolution in synergy with the thermodynamic evolution. By operating close to maximal entropy production, photoconverters couple their own (biological) evolution to thermodynamic evolution in a positive feedback loop which speeds up both evolutions. This “evolution coupling” hypothesis postulates that biosphere evolution is intimately connected with the evolution of life’s physical environment as suggested by the Gaia hypothesis. In this picture biological evolution is just a clever way nature found to accelerate its thermodynamic evolution. Life’s particular goal seems to be to channel the input power into those dissipative pathways where electrochemical rather than only thermal free-energy conversions can occur (Juretić and Županović, 2005).”
To be more specific, our group in Split pointed out that linear nonequilibrium thermodynamics can be derived from the MEP principle. In other words there is an equivalency between Onsager principle of the least dissipation of energy and the MEP principle in this simple case close to thermodynamic equilibrium (Županović et al., FIZIKA A 14 (2005) 89–96, and Entropy 12 (2010) 996–1005.). Also we proved that Kirchhoff's voltage law can be derived from two more general principles: MEP and energy conservation (Županović and Juretić, Phys. Rev. E 70, 056108 (2004)).
Is there any quantitative evidence that MEP (MaxEP) principle is predictive in bioenergetics and enzyme kinetics, either close or far from thermodynamic equilibrium? Indeed, we found such evidence in photosynthesis, ATP-synthase molecular motor, beta-lactamase, and triosephosphate isomerase (TIM). We used MEP and/or MaxENT principle to predict optimal values of kinetic constants for simplified kinetic models and got sometimes very good (ATP-synthase) and more often only rough agreement with experimental values of kinetic constants. It is worth pointing out that minimal entropy production principle by I. Prigogine cannot help at all for predicting rate constants or catalytic constants.
Our latest paper (Bonačić Lošić, Donđivić and Juretić 2016) went a step further and solely by using the MEP principle accomplished several important goals:
- It proved that triosephosphate isomerase (TIM) is not a fully evolved enzyme with near-maximal possible reaction rate, as often stated by biochemists
- It identified product (R-glyceraldehyde-3-phosphate) release as the rate-limiting step
- It proved that only the MaxEP requirement for the product release step led to 30% increase in enzyme activity, specificity constant kcat/KM, and overall entropy production
- It connected MEP principle with kinetic and structural studies opening the possibility to find amino acid substitutions leading to an increased frequency of loop six opening and product release
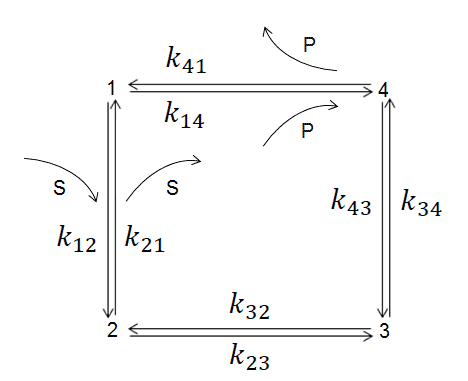
The four-state kinetic diagram for TIM enzyme, where kij and kji are transition kinetic constants (i= 1, 2, 3, 4). Functional states are: 1 the enzyme (E), 2 the enzyme-substratecomplex (ES), 3 an intermediate (EZ), and 4 the enzyme-product complex (EP)
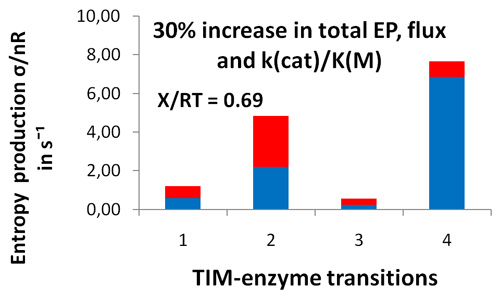
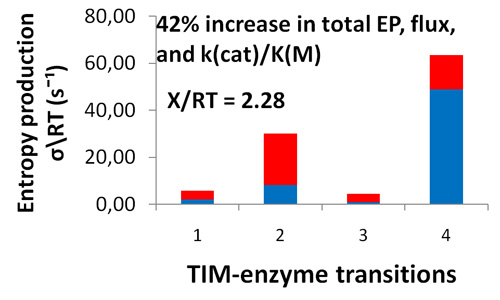
Distribution of entropy productions for each of four TIM-enzyme functional transitions before (blue columns) and after (extended red area over blue area in each column) MaxEP optimization for the 4-th step (product release). Product (GAP) concentration was 0.064 μM (upper figure) or 0.013 μM (lower figure).
Secondary structure prediction for membrane proteins
Now classic Juretić et al. paper 2002 collected a large number of citations (195), because high quality predictions for proteins segments with preference to enter membrane as shorter or longer (transmembrane) helices are still very valuable. Versions SPLIT 3.5 and SPLIT 4 are available for online scientific calculations and have been valuable free bioinformatic resources for more than 15 years. An earlier version of the SPLIT algorithm was the first online server for scientific calculations in Croatia. Better versions of SPLIT algorithm can be constructed today by using much higher number of integral membrane proteins with known sequence location of transmembrane helices during the training procedure for extracting preference functions, and by eliminating known weaknesses such as:
- too high percentage of false TMH predictions in soluble proteins,
- too high false TMH prediction for signal sequences,
- neglecting homologous sequences to the tested one.